文献速递:生成对抗网络医学影像中的应用——3DGAUnet:一种带有基于3D U-Net的生成器的3D生成对抗网络
文献速递:生成对抗网络医学影像中的应用——3DGAUnet:一种带有基于3D U-Net的生成器的3D生成对抗网络
给大家分享文献的主题是生成对抗网络(Generative adversarial networks, GANs)在医学影像中的应用。文献的研究内容包括同模态影像生成、跨模态影像生成、GAN在分类和分割方面的应用等。生成对抗网络与其他方法相比展示出了优越的数据生成能力,使它们在医学图像应用中广受欢迎。这些特性引起了医学成像领域研究人员的浓厚兴趣,导致这些技术在各种传统和新颖应用中迅速实施,如图像重建、分割、检测、分类和跨模态合成。
01
文献速递介绍
胰腺导管腺癌(PDAC)代表了一个重大的公共卫生问题,这是由于其诊断延迟、当前化疗治疗的有限效果以及总体预后不佳。它在所有主要实体恶性肿瘤中具有最高的致死率。尽管几十年来在临床和研究上做了大量努力,但一年生存率仍然只有20%,而五年生存率在相当长的时间内一直处于个位数,直到最近才提高到11%。尽管如果在局部阶段实现早期检测,5年相对生存率有潜力大幅提高至42% ,但目前缺乏可靠识别早期无症状胰腺癌的明确筛查方法。
计算机断层扫描(CT)是主要的诊断成像方法之一。近年来,基于深度学习的方法越来越被视为多功能应用。它们可以将物理和语义细节直接整合到神经网络架构中,并被用于解决医学成像中的计算机视觉任务,如胸部X光片和组织病理学图像的分割、配准和分类 。例如,卷积神经网络(CNN)模型在自然图像和医学图像的图像分类任务中表现出高可行性,从2D模型到3D模型。一些类似的研究已经应用于胰腺癌分类器,用于分析和解释医学成像数据中的特征 。
在开发用于图像任务的深度学习模型时,通常需要大量的数据集(例如,成千上万的图像),以确保模型在不过拟合的情况下收敛。然而,尤其是对于PDAC,临床信息的可用性经常受到队列规模小的限制,这给实现最佳模型训练带来了障碍。研究人员已经开发了如数据增强、生成对抗网络(GAN)、交叉验证和优化方法(如锐化感知最小化 )等方法,以克服训练数据的不足。生成模型在医学图像合成方面显示出了效果,特别是在2D成像模式下。最近,研究人员已经开发了基于2D的GAN模型,用于生成胰腺肿瘤的逼真CT图像 。然而,在PDAC的背景下使用3D生成模型仍然受到限制,直接应用现有方法(例如,3D-GAN)可能无法实现合成特定于PDAC的三维CT图像数据的理想结果。PDAC肿瘤通常表现出微妙的成像特征,因为它们可能与周围的胰腺组织同密度或低密度,使得它们在视觉上难以区分。此外,PDAC肿瘤可能缺乏明确的边界,使得它们与正常的胰腺实质区分开来具有挑战性。因此,开发有效的技术来增强3D PDAC肿瘤数据集至关重要,以促进深度学习模型在解决PDAC方面的进展。
我们开发了一种基于GAN的工具,能够生成逼真的3D CT图像,展现PDAC肿瘤和胰腺组织。为了克服这些挑战并使这个3D GAN模型表现更好,我们的创新之处在于为生成器开发了一个3D U-Net架构,以改善PDAC肿瘤和胰腺组织的形状和纹理学习。3D U-Net在医学图片自动分割中的应用显示了适当和优越的结果。值得注意的是,这是它首次被整合到GAN模型中。这个3D GAN模型分别生成PDAC肿瘤组织CT图像和健康胰腺组织CT图像的体积数据,并采用了一种混合方法来创建逼真的最终图像。对开发的3D GAN模型进行了多个数据集的全面检查和验证,以确定模型在临床环境中的有效性和适用性。我们通过使用合成图像数据训练3D CNN模型来预测3D肿瘤斑块,评估了我们方法的有效性。开发了一个软件包3DGAUnet,用于实现这个带有3D U-Net基础生成器的3D生成对抗网络,用于肿瘤CT图像合成。这个软件包有可能被适配到其他类型的实体肿瘤,因此在图像处理模型方面为医学成像领域做出了重大贡献。这个软件包可在https://github.com/yshi20/3DGAUnet上获取(2023年10月12日访问)。
Title
题目
3DGAUnet: 3D Generative Adversarial Networks with a 3D U-Net Based Generator to Achieve the Accurate and Effective Synthesis of Clinical Tumor Image Data for Pancreatic Cancer
3DGAUnet:一种带有基于3D U-Net的生成器的3D生成对抗网络,用于实现胰腺癌临床肿瘤图像数据的精确和有效合成。
Abstract
摘要
Pancreatic ductal adenocarcinoma (PDAC) presents a critical global health challenge, andearly detection is crucial for improving the 5-year survival rate. Recent medical imaging and computational algorithm advances offer potential solutions for early diagnosis. Deep learning, particularly in the form of convolutional neural networks (CNNs), has demonstrated success in medical image analysis tasks, including classification and segmentation. However, the limited availability of clinicaldata for training purposes continues to represent a significant obstacle. Data augmentation, gen**erative adversarial networks (GANs), and cross-validation are potential techniques to address this limitation and improve model performance, but effective solutions are still rare for 3D PDAC, where the contrast is especially poor, owing to the high heterogeneity in both tumor and background tissues. In this study, we developed a new GAN-based model, named 3DGAUnet, for generating realistic 3D CT images of PDAC tumors and pancreatic tissue, which can generate the inter-slice connection
data that the existing 2D CT image synthesis models lack. The transition to 3D models allowed the preservation of contextual information from adjacent slices, improving efficiency and accuracy,
especially for the poor-contrast challenging case of PDAC. PDAC’s challenging characteristics, such as an iso-attenuating or hypodense appearance and lack of well-defined margins, make tumor shape
and texture learning challenging. To overcome these challenges and improve the performance of 3D GAN models, our innovation was to develop a 3D U-Net architecture for the generator, to improve
shape and texture learning for PDAC tumors and pancreatic tissue. Thorough examination and validation across many datasets were conducted on the developed 3D GAN model, to ascertain
the efficacy and applicability of the model in clinical contexts. Our approach offers a promising path for tackling the urgent requirement for creative and synergistic methods to combat PDAC. The development of this GAN-based model has the potential to alleviate data scarcity issues, elevate the quality of synthesized data, and thereby facilitate the progression of deep learning models, to enhance the accuracy and early detection of PDAC tumors, which could profoundly impact patient outcomes. Furthermore, the model has the potential to be adapted to other types of solid tumors, hence making significant contributions to the field of medical imaging in terms of image processing models.
胰腺导管腺癌(PDAC)呈现出一个严峻的全球健康挑战,早期检测对于提高5年生存率至关重要。近期医学影像和计算算法的进步为早期诊断提供了潜在解决方案。深度学习,特别是卷积神经网络(CNNs)的形式,在医学图像分析任务中,包括分类和分割,已经展现出成功。然而,临床数据用于训练目的的有限可用性仍然代表着一个重大障碍。数据增强、生成对抗网络(GANs)和交叉验证是解决这一限制并改善模型性能的潜在技术,但对于3D PDAC特别是对比度特别差的情况,由于肿瘤和背景组织的高度异质性,有效解决方案仍然很少。
在这项研究中,我们开发了一种新的基于GAN的模型,名为3DGAUnet,用于生成现实的PDAC肿瘤和胰腺组织的3D CT图像,这些图像可以生成现有2D CT图像合成模型缺乏的片间连接数据。转向3D模型允许从相邻切片保留上下文信息,提高效率和准确性,特别是对于对比度差的PDAC挑战性病例。PDAC的挑战性特征,如等密度或低密度外观和缺乏明确边界,使得肿瘤形状和纹理学习具有挑战性。为了克服这些挑战并提高3D GAN模型的性能,我们的创新之处是开发了一个用于生成器的3D U-Net架构,以改善PDAC肿瘤和胰腺组织的形状和纹理学习。对开发的3D GAN模型进行了彻底的检查和验证,以确定该模型在临床环境中的有效性和适用性。我们的方法为应对PDAC紧急需求的创新和协同方法提供了一条有希望的途径。这种基于GAN的模型的开发有可能减轻数据稀缺问题,提高合成数据质量,并从而促进深度学习模型的发展,以提高PDAC肿瘤的准确性和早期检测,这可能对患者结果产生深远影响。
此外,该模型有潜力适应于其他类型的实体肿瘤,因此在医学影像领域的图像处理模型方面做出重大贡献。
Methods
方法
Figure 1a illustrates the overall workflow of our proposed method. Given a set of PDAC CT images that can be acquired through different sources, we first conduct data preprocessing on these raw image data to tackle data heterogeneity and generate normalized and resampled volume data for tumor tissues and pancreas. These preprocessed
datasets are then used as the training set and fed into 3DGAUnet, the new 3D GAN model developed in this work for tumor CT image synthesis. After the tumor and pancreastypes are learned independently via 3DGAUnet, the corresponding synthetic data can be generated.To effectively combine these synthetic tissues, we evaluated three blending methods and identified the most suitable technique for PDAC tumor CT images. Given that the pancreas is a parenchymal organ, the relative location of the tumor tissue was found to be less significant. As a result, the focus was primarily on blending the different tissue types seamlessly and realistically to ensure accurate and reliable results for diagnosing PDAC tumors in CT images.We evaluated the usability of the synthetic data by applying it in a diagnosis task.
For this purpose, we employed a 3D CNN classifier capable of taking 3D volumes as input, which was an improvement over the traditional classification tools that only use individual**slices and overlook the inter-slice information.
By integrating the synthetic data, we addressed common challenges encountered in real-world scenarios, such as the small size of the dataset and imbalanced data. The addition of synthetic samples helped to improve the model’s performance and mitigate issues related to imbalanced datasets.
图1a阐述了我们提出方法的整体工作流程。给定一组可以通过不同来源获得的胰腺导管腺癌(PDAC)CT图像,我们首先对这些原始图像数据进行数据预处理,以解决数据异质性问题,并生成肿瘤组织和胰腺的标准化和重采样的体积数据。这些预处理后的数据集随后被用作训练集,并输入到3DGAUnet,这是我们在这项工作中开发的用于肿瘤CT图像合成的新3D GAN模型。通过3DGAUnet分别学习了肿瘤和胰腺类型后,可以生成相应的合成数据。
为了有效地结合这些合成组织,我们评估了三种混合方法,并确定了用于PDAC肿瘤CT图像的最合适技术。鉴于胰腺是实质器官,肿瘤组织的相对位置被发现不太重要。因此,重点主要放在无缝且现实地混合不同组织类型上,以确保准确和可靠的PDAC肿瘤CT图像诊断结果。
我们通过将其应用于诊断任务来评估合成数据的可用性。为此,我们使用了一个能够接受3D体积作为输入的3D CNN分类器,这比仅使用单个切片并忽略片间信息的传统分类工具有所改进。
通过整合合成数据,我们解决了现实世界场景中常见的挑战,例如数据集的小规模和数据不平衡。合成样本的添加有助于提高模型性能,减轻与数据不平衡有关的问题。
Results
结果
We trained our 3DGAUnet model separately using PDAC tumor and healthy pancreas data. These are referred to as the tumor and pancreas models. The tumor model was trained using PDAC tumor data, including 174 volumetric tumor data in Nifty format.
The pancreas model was trained using healthy tissue data, including 200 volumetric data in Nifty format. Both input datasets resulted from the preprocessing steps outlined in Section 2.1. Image augmentation, including image flipping and rotation, was performed on the training data. The augmented data for each volume were generated by rotating each volume on three axes in 12? , 24? , 36? , 48? , and 72? increments. All images for the tumor model were resampled to 1 mm isotropic resolution and trimmed to 32 × 32 × 32 size.All images for the pancreas model were resampled to 1 mm isotropic resolution and trimmed to 64 × 64 × 64 size, with pancreas tissue filling the entire cube. The training procedure of any GAN model is inherently unstable because of the dynamic of optimizing two competing losses. For each model in this study, the training process saved the model weights every 20 epochs, and the entire model was trained for 2000 epochs. The best training duration before the model collapsed was decided by inspecting the generator loss curve and finding the epoch before the loss drastically increased. We trained our models with an NVIDIA RTX 3090 GPU. The optimal parameter set was searched within a parameter space consisting of batch size and learning rate, where the possible batch sizes included 4, 8, 16, and 32, and the possible learning rates included**0.1, 0.01, 0.001, 0.0001, and 0.00001.
A set of 500 synthetic volumetric data were generated by the tumor and pancreas models separately. We first conducted a qualitative comparison between the training image sets and the synthetic image sets. We used volume rendering to visualize these datasets, to inspect the 3D results. Figure 2a shows examples of ground truth tumor volumes, synthetic tumors generated by the existing technique 3D-GAN [18], and synthetic tumors generated by our 3DGAUnet. We can see that when we trained our 3DGAUnet based on the group truth inputs, our model could generate synthetic tumors carrying realistic an anatomical structure and texture and capturing overall shape and details. Nonetheless, 3D-GAN either produced unsuccessful data or failed to generate meaningful results in capturing the tumor’s geometry. Figure 2b shows examples of the group truth pancreas volumes and
synthetic pancreas volumes generated by 3D-GAN and our 3DGAUnet. By comparing the generated pancreas volumes with real medical images, we can see that our 3DGUnet could effectively synthesize a 3D pancreas to resemble actual anatomical structures. However, it was hard for 3D-GAN to generate anatomically plausible results, and a certain ambient noise was perceived in the generated volumes. We further examined the interior structures of the volumes generated by our 3DGUnet. Figure 3 shows the 2D slices of the ground truth, 3D-GAN, and our 3DGAUnet images from both tumor and pancreas models. The synthetic data produced by our 3DGAUnet model exhibited a high degree of fidelity to the ground truth, in terms of both internal anatomical structure and texture, compared to 3D-GAN. In certain instances,
我们分别使用PDAC肿瘤和健康胰腺数据训练了我们的3DGAUnet模型。这些被称为肿瘤和胰腺模型。肿瘤模型是使用包括174个Nifty格式的体积肿瘤数据的PDAC肿瘤数据训练的。胰腺模型是使用包括200个Nifty格式的体积数据的健康组织数据训练的。两个输入数据集都是根据第2.1节中概述的预处理步骤生成的。对训练数据进行了图像增强,包括图像翻转和旋转。通过在三个轴上以12°、24°、36°、48°和72°的增量旋转每个体积来生成每个体积的增强数据。所有肿瘤模型的图像都被重新采样为1 mm等距分辨率,并裁剪为32×32×32大小。所有胰腺模型的图像都被重新采样为1 mm等距分辨率,并裁剪为64×64×64大小,其中胰腺组织填满了整个立方体。
任何GAN模型的训练过程本质上是不稳定的,因为它涉及优化两个相互竞争的损失。在本研究中,每个模型的训练过程每20个周期保存一次模型权重,整个模型训练了2000个周期。通过检查生成器损失曲线并找到损失急剧增加之前的周期,确定了模型崩溃前的最佳训练持续时间。我们使用NVIDIA RTX 3090 GPU训练了我们的模型。最佳参数集是在由批量大小和学习率组成的参数空间中搜索得到的,其中可能的批量大小包括4、8、16和32,可能的学习率包括0.1、0.01、0.001、0.0001和0.00001。
肿瘤和胰腺模型分别生成了一组500个合成的体积数据。我们首先对训练图像集和合成图像集进行了定性比较。我们使用体积渲染来可视化这些数据集,以检查3D结果。图2a展示了真实肿瘤体积、现有技术3D-GAN [18]生成的合成肿瘤和我们的3DGAUnet生成的合成肿瘤的例子。我们可以看到,当我们基于真实输入训练我们的3DGAUnet时,我们的模型可以生成具有逼真的解剖结构和纹理的合成肿瘤,并捕获整体形状和细节。然而,3D-GAN要么产生了失败的数据,要么无法在捕获肿瘤的几何形状方面产生有意义的结果。图2b展示了真实胰腺体积和由3D-GAN及我们的3DGAUnet生成的合成胰腺体积的例子。通过将生成的胰腺体积与真实医学图像进行比较,我们可以看到我们的3DGUnet能够有效地合成类似于实际解剖结构的3D胰腺。然而,3D-GAN难以生成解剖学上可信的结果,并且在生成的体积中感知到了一定的环境噪音。
Conclusions
结论
The 3DGAUnet model represents a significant advancement in synthesizing clinical tumor CT images, providing realistic and spatially coherent 3D data, and it holds great potential for improving medical image analysis and diagnosis. In the future, we will
continue to address problems associated with the topic of computer vision and cancer. We would like to investigate reasonable usages of synthetic data and evaluations of data quality and usability in practice; for example, their effectiveness in training reliable classification models. We also plan to conduct reader studies involving domain experts (e.g., radiologists), to assess whether synthetic CTs can enhance diagnostic accuracy. This would offeradditional clinical validation regarding the resemblance of synthesized CTs to real-world**PDAC tumor characteristics.
3DGAUnet模型在合成临床肿瘤CT图像方面代表了重大进步,提供了现实且空间连贯的3D数据,并且对改善医学图像分析和诊断具有巨大潜力。未来,我们将继续解决与计算机视觉和癌症相关的问题。我们希望研究合成数据的合理用途以及实践中数据质量和可用性的评估;例如,它们在训练可靠的分类模型中的有效性。我们还计划进行涉及领域专家(例如,放射科医师)的读者研究,以评估合成CT是否可以提高诊断准确性。这将提供关于合成CT与现实世界胰腺导管腺癌(PDAC)肿瘤特征相似性的额外临床验证。
Figure
图
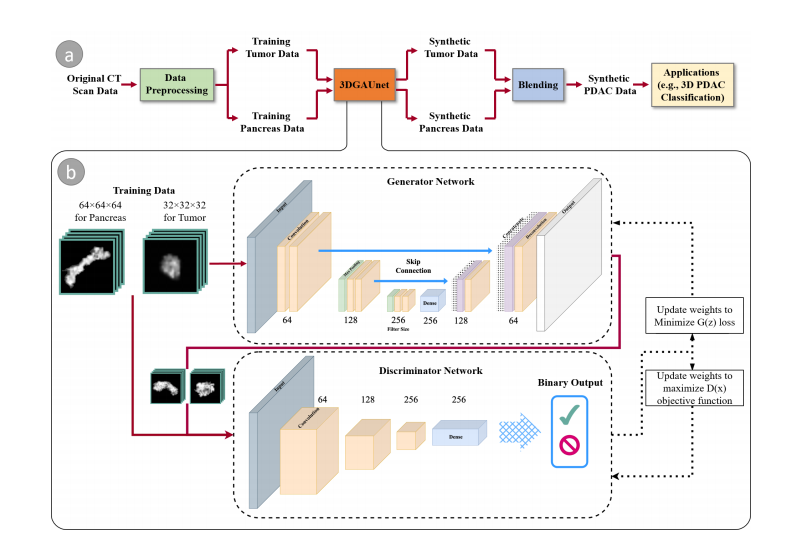
Figure 1. An overview of our method. (a) the workflow components and (b) the architecture of our GAN-based model, 3DGAUnet, consisting of a 3D U-Net-based generator network and a 3D CNN-based discriminator network to generate synthetic data.
图1. 我们方法的概览。(a)工作流程组件和(b)基于GAN的模型3DGAUnet的架构,包括一个基于3D U-Net的生成器网络和一个基于3D CNN的判别器网络,用于生成合成数据。
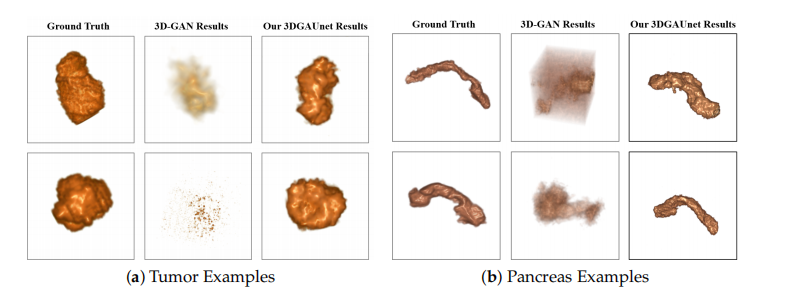
Figure 2. Examples of 3D volumes obtained from the different methods. Examples of 3D volume data of tumor (a) and pancreas (b) from the different methods. In each set of examples, the left, middle, and right columns correspond to ground truth data, synthetic data generated by 3D-GAN, and synthetic data generated by our 3DGAUnet, respectively. All 3D volumes are shown in vol ume rendering.
图2. 从不同方法获得的3D体积示例。来自不同方法的肿瘤(a)和胰腺(b)的3D体积数据示例。在每组示例中,左列、中列和右列分别对应于真实数据、由3D-GAN生成的合成数据和由我们的3DGAUnet生成的合成数据。所有3D体积均以体积渲染方式显示。
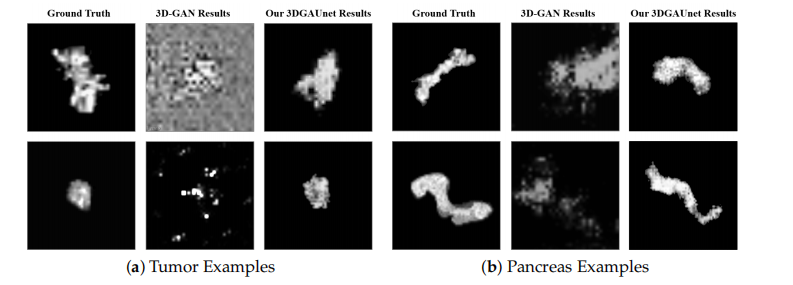
Figure 3. Examples of 2D slices in 3D volumes obtained from different methods. Examples of 2D *in 3D volumes of tumor (a) and pancreas (b) from the different resources. In each set of examples,*the left, middle, and right columns correspond to ground truth data, synthetic data generated by 3D-GAN, and synthetic data generated by our 3DGAUnet, respectively.
图3. 从不同方法获得的3D体积中2D切片的示例。来自不同资源的肿瘤(a)和胰腺(b)在3D体积中的2D切片示例。在每组示例中,左列、中列和右列分别对应于真实数据、由3D-GAN生成的合成数据和由我们的3DGAUnet生成的合成数据。

Figure 4. Comparison of the blending methods. For a ground truth PDAC tissue data (a), the image (b) provides a close-up view of the texture at the tumor site within the red box in (a), serving as a**visual reference. The images (c–e) show the blend of a tumor into the healthy pancreas tissue using different blending methods. We can observe that Blend III had the best visual similarity
图4. 混合方法的比较。对于真实的胰腺导管腺癌(PDAC)组织数据(a),图像(b)提供了红色框中(a)肿瘤部位的纹理的特写视图,作为视觉参考。图像(c–e)显示了使用不同混合方法将肿瘤融入健康胰腺组织。我们可以观察到,Blend III具有最佳的视觉相似性。
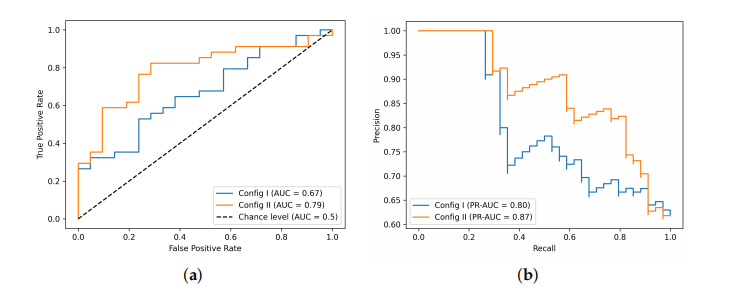
Figure 5. 3D CNN classifier performance. (a) ROC curves and (b) PR curves with two configurations of training datasets.
图5. 3D CNN分类器性能。(a)ROC曲线和(b)两种训练数据集配置的PR曲线。
Table
表
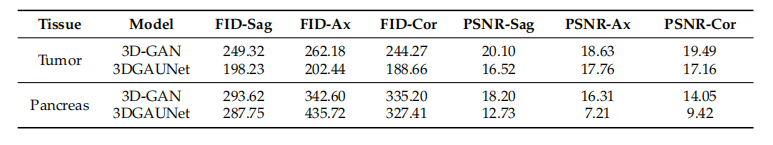
Table 1. Performance based on 2D image quality metrics.
表1. 基于2D图像质量指标的性能。
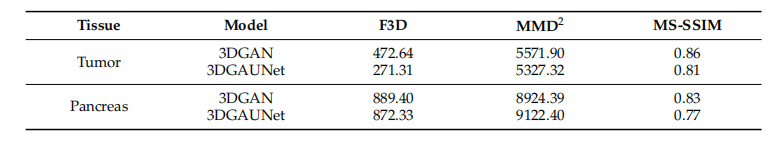
Table 2. Performance based on 3D image quality metrics.
表2. 基于3D图像质量指标的性能。
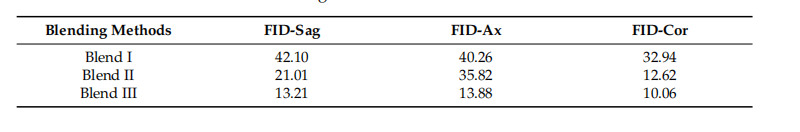
Table 3. Slice-wise FID values of blending methods.
表3. 混合方法的逐片FID值。
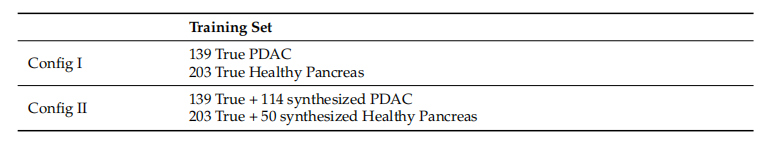
Table 4. Dataset configurations for classifier experiments.
表4. 分类器实验的数据集配置。
本文来自互联网用户投稿,该文观点仅代表作者本人,不代表本站立场。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。 如若内容造成侵权/违法违规/事实不符,请联系我的编程经验分享网邮箱:veading@qq.com进行投诉反馈,一经查实,立即删除!