Epicypher:CUTANA? E. coli Spike-in DNA
来源于Escherichia coli(E.coli)的片段DNA可以用作核酸酶靶向切割和释放(CUT&RUN)的实验标准化的spike-in对照。产品CUTANA? E. coli Spike-in DNA含有足够的材料,可用于100-200个CUT&RUN样本(高丰度和低丰度靶标的范围)。
产品详情
保存温度:?Stable for 2 years at -20°C from date of receipt. After resuspending, aliquots should be stored at -20°C.
运输温度:?Room temp. Can go on frozen cold packs or dry ice.
产品形式:?100 ng lyophilized DNA. *NOTE: May not be visible.
验证数据
|
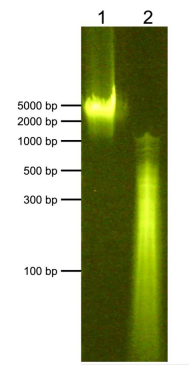
| Figure 1: DNA gel data E. coli fragment size distribution. Lane 1: gDNA extracted from JM101 E.? coli cells (500 ng). Lane 2: Digested and purified CUTANA? E. coli Spike-in? DNA (500 ng) resolved via 2% E-Gel? EX Agarose Gel. Migration positions? of DNA molecular weight markers are indicated. |
|
|
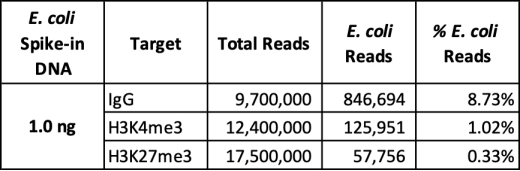
| Figure 2: CUT&RUN sequencing data CUTANA? E. coli Spike-in DNA (1.0 ng) was added to CUT&RUN samples using 500,000 K562 cells enriched? for a low abundance target (H3K4me3, EpiCypher 13-0041), a high abundance target (H3K27me3, EpiCypher? 13-0030) and an IgG negative control (EpiCypher 13-0042). Total numbers of paired-end sequencing reads, reads? aligned to E. coli, and percentage of total sequencing reads aligned to E. coli spike-in DNA are shown. NOTE:? Spike-in DNA amount should be optimized by the end user with the goal of E. coli DNA comprising ~1% (0.2 -? 5%) of total sequencing reads. |
产品详情
| 货号 | 产品名称 | 规格 |
| 18-1401 | CUTANA? E. coli Spike-in DNA / 欣博盛生物 | 100 ng |
本文来自互联网用户投稿,该文观点仅代表作者本人,不代表本站立场。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。 如若内容造成侵权/违法违规/事实不符,请联系我的编程经验分享网邮箱:veading@qq.com进行投诉反馈,一经查实,立即删除!