evo_ape tum指令使用及各参数使用效果
目录
evo_ape tum gt.txt l.txt -a -p?
evo_ape tum gt.txt l.txt -s -p
evo_ape tum gt.txt l.txt -as -p
evo_ape tum gt.txt l.txt? --n_to_align 1
evo_ape tum gt.txt l.txt -as -p --plot_mode=xy
evo_ape tum gt.txt l.txt -as -p? --plot_colormap_max 0.2
evo_ape tum gt.txt l.txt -as -p --plot_colormap_min 0.2
evo_ape tum gt.txt l.txt -as -p --plot_colormap_max_percentile 50
evo_ape tum gt.txt l.txt -as -p? --plot_full_ref
evo_ape tum gt.txt l.txt -as? --save_plot SAVE_PLOT.pdf
evo_ape tum gt.txt l.txt -as --serialize_plot SERIALIZE_PLOT
evo_ape tum gt.txt l.txt -as --save_results ape_result.zip
evo_ape tum gt.txt l.txt -as --logfile ape_result.log
evo_ape tum gt.txt l.txt -as? --no_warnings
evo_ape tum gt.txt l.txt -as --silent
?evo_ape tum gt.txt l.txt -as?? --debug
学习evo_ape tum指令的使用
参数含义
查看帮助
evo_ape tum -h
$ evo_ape tum -h
usage: evo_ape tum [-h] [-r {full,trans_part,rot_part,angle_deg,angle_rad}]
?????????????????? [-a] [-s] [--n_to_align N_TO_ALIGN] [--align_origin] [-p]
?????????????????? [--plot_mode {xy,xz,yx,yz,zx,zy,xyz}]
?????????????????? [--plot_colormap_max PLOT_COLORMAP_MAX]
?????????????????? [--plot_colormap_min PLOT_COLORMAP_MIN]
?????????????????? [--plot_colormap_max_percentile PLOT_COLORMAP_MAX_PERCENTILE]
?????????????????? [--plot_full_ref] [--ros_map_yaml ROS_MAP_YAML]
?????????????????? [--save_plot SAVE_PLOT] [--serialize_plot SERIALIZE_PLOT]
?????????????????? [--save_results SAVE_RESULTS] [--logfile LOGFILE]
?????????????????? [--no_warnings] [-v] [--silent] [--debug] [-c CONFIG]
?????????????????? [--t_max_diff T_MAX_DIFF] [--t_offset T_OFFSET]
?????????????????? ref_file est_fileAbsolute pose error (APE) metric app for TUM trajectory files - (c) evo
authorspositional arguments:
? ref_file????????????? reference trajectory file
? est_file????????????? estimated trajectory fileoptional arguments:
? -h, --help??????????? show this help message and exit
? --t_max_diff T_MAX_DIFF
??????????????????????? maximum timestamp difference for data association
? --t_offset T_OFFSET?? constant timestamp offset for data associationalgorithm options:
? -r {full,trans_part,rot_part,angle_deg,angle_rad}, --pose_relation {full,trans_part,rot_part,angle_deg,angle_rad}
??????????????????????? pose relation on which the APE is based
? -a, --align?????????? alignment with Umeyama's method (no scale)
? -s, --correct_scale?? correct scale with Umeyama's method
? --n_to_align N_TO_ALIGN
??????????????????????? the number of poses to use for Umeyama alignment,
??????????????????????? counted from the start (default: all)
? --align_origin??????? align the trajectory origin to the origin of the
??????????????????????? reference trajectoryoutput options:
? -p, --plot??????????? show plot window
? --plot_mode {xy,xz,yx,yz,zx,zy,xyz}
??????????????????????? the axes for plot projection
? --plot_colormap_max PLOT_COLORMAP_MAX
??????????????????????? the upper bound used for the color map plot (default:
??????????????????????? maximum error value)
? --plot_colormap_min PLOT_COLORMAP_MIN
??????????????????????? the lower bound used for the color map plot (default:
??????????????????????? minimum error value)
? --plot_colormap_max_percentile PLOT_COLORMAP_MAX_PERCENTILE
??????????????????????? percentile of the error distribution to be used as the
??????????????????????? upper bound of the color map plot (in %, overrides
??????????????????????? --plot_colormap_max)
? --plot_full_ref?????? plot the full, unsynchronized reference trajectory
? --ros_map_yaml ROS_MAP_YAML
??????????????????????? yaml file of an ROS 2D map image (.pgm/.png) that will
??????????????????????? be drawn into the plot
? --save_plot SAVE_PLOT
??????????????????????? path to save plot
? --serialize_plot SERIALIZE_PLOT
??????????????????????? path to serialize plot (experimental)
? --save_results SAVE_RESULTS
??????????????????????? .zip file path to store results
? --logfile LOGFILE???? Local logfile path.usability options:
? --no_warnings???????? no warnings requiring user confirmation
? -v, --verbose???????? verbose output
? --silent????????????? don't print any output
? --debug?????????????? verbose output with additional debug info
? -c CONFIG, --config CONFIG
??????????????????????? .json file with parameters (priority over command line
??????????????????????? args)
翻译之后
$ evo_ape tum -h
usage: evo_ape tum [-h] [-r {full,trans_part,rot_part,angle_deg,angle_rad}]
?????????????????? [-a] [-s] [--n_to_align N_TO_ALIGN] [--align_origin] [-p]
?????????????????? [--plot_mode {xy,xz,yx,yz,zx,zy,xyz}]
?????????????????? [--plot_colormap_max PLOT_COLORMAP_MAX]
?????????????????? [--plot_colormap_min PLOT_COLORMAP_MIN]
?????????????????? [--plot_colormap_max_percentile PLOT_COLORMAP_MAX_PERCENTILE]
?????????????????? [--plot_full_ref] [--ros_map_yaml ROS_MAP_YAML]
?????????????????? [--save_plot SAVE_PLOT] [--serialize_plot SERIALIZE_PLOT]
?????????????????? [--save_results SAVE_RESULTS] [--logfile LOGFILE]
?????????????????? [--no_warnings] [-v] [--silent] [--debug] [-c CONFIG]
?????????????????? [--t_max_diff T_MAX_DIFF] [--t_offset T_OFFSET]
?????????????????? ref_file est_file
Absolute pose error (APE) metric app for TUM trajectory files - (c) evo
authors
TUM轨迹文件的绝对姿态误差(APE)度量应用程序-(c)evo
作者
positional arguments:
位置参数:
? ref_file????????????? reference trajectory file参考轨迹文件
? est_file????????????? estimated trajectory file估计轨迹文件
optional arguments:可选参数:
? -h, --help??????????? show this help message and exit显示此帮助消息并退出
? --t_max_diff T_MAX_DIFF
??????????????????????? maximum timestamp difference for data association数据关联的最大时间戳差异
? --t_offset T_OFFSET?? constant timestamp offset for data association数据关联的常量时间戳偏移量
algorithm options:
算法选项:
? -r {full,trans_part,rot_part,angle_deg,angle_rad}, --pose_relation {full,trans_part,rot_part,angle_deg,angle_rad}
????????????????????? ? ?? pose relation on which the APE is basedAPE所基于的姿态关系
? -a, --align?????????? alignment with Umeyama's method (no scale)与Umeyama的方法对齐(无比例)
? -s, --correct_scale?? correct scale with Umeyama's method用Umeyama的方法校正刻度
? --n_to_align N_TO_ALIGN
??????????????????????? the number of poses to use for Umeyama alignment,counted from the start (default: all)用于Umeyama对齐的姿势数,从开始计数(默认值:全部)
? --align_origin??????? align the trajectory origin to the origin of the reference trajectory将轨迹原点与参考轨迹的原点对齐
output options:输出选项:
? -p, --plot??????????? show plot window显示绘图窗口
? --plot_mode {xy,xz,yx,yz,zx,zy,xyz}
??????????????????????? the axes for plot projection绘图投影的轴
? --plot_colormap_max PLOT_COLORMAP_MAX
??????????????????????? the upper bound used for the color map plot (default: maximum error value)用于颜色映射图的上限(默认值:最大误差值)
? --plot_colormap_min PLOT_COLORMAP_MIN
??????????????????????? the lower bound used for the color map plot (default:minimum error value)用于颜色映射图的下限(默认值:最小误差值)
? --plot_colormap_max_percentile PLOT_COLORMAP_MAX_PERCENTILE
??????????????????????? percentile of the error distribution to be used as the? upper bound of the color map plot (in %, overrides? --plot_colormap_max)要用作颜色映射图上限的错误分布的百分位数(以%为单位,覆盖--plot_colormap_max)
? --plot_full_ref?????? plot the full, unsynchronized reference trajectory绘制完整的、不同步的参考轨迹
? --ros_map_yaml ROS_MAP_YAML
??????????????????????? yaml file of an ROS 2D map image (.pgm/.png) that will be drawn into the plot
? --save_plot SAVE_PLOT
??????????????????????? path to save plot保存绘图的路径
? --serialize_plot SERIALIZE_PLOT
??????????????????????? path to serialize plot (experimental)序列化绘图的路径(实验)
? --save_results SAVE_RESULTS
??????????????????????? .zip file path to store results存储结果的.zip文件路径
? --logfile LOGFILE???? Local logfile path.本地日志文件路径。
usability options:可用性选项:
? --no_warnings???????? no warnings requiring user confirmation无需用户确认的警告
? -v, --verbose???????? verbose output详细输出
? --silent????????????? don't print any output不打印任何输出
? --debug?????????????? verbose output with additional debug info使用附加调试信息调试详细输出
? -c CONFIG, --config CONFIG
??????????????????????? .json file with parameters (priority over command line args)带有参数的.json文件(优先级高于命令行args)
效果
受到数据影响,很多参数的效果无法有效展示。
evo_ape tum gt.txt l.txt
$ evo_ape tum gt.txt l.txt
APE w.r.t. translation part (m)
(not aligned)?????? max?? ?6372512.914678
????? mean?? ?6372511.473926
??? median?? ?6372511.871822
?????? min?? ?6372508.722174
????? rmse?? ?6372511.473926
?????? sse?? ?934004757162323.000000
?????? std?? ?1.138090
?算法选项
--t_max_diff T_MAX_DIFF
??????????????????????? maximum timestamp difference for data association数据关联的最大时间戳差异
? --t_offset T_OFFSET?? constant timestamp offset for data association数据关联的常量时间戳偏移量
如果两个txt的实际时间戳存在固定差值m,可以用“? --t_offset? m”
evo_ape tum gt.txt l.txt -a -p?
? -a, --align?????????? alignment with Umeyama's method (no scale)用Umeyama的方法对齐(无比例)
$ evo_apeum gt.txt l.txt -a -p
APE w.r.t. translation part (m)
(with SE(3) Umeyama alignment)?????? max?? ?0.869683
????? mean?? ?0.252346
??? median?? ?0.222082
?????? min?? ?0.024169
????? rmse?? ?0.330618
?????? sse?? ?2.514097
?????? std?? ?0.213612


evo_ape tum gt.txt l.txt -s -p
$ evo_ape tum gt.txt l.txt -s -p
APE w.r.t. translation part (m)
(scale corrected)?????? max?? ?6372512.113874
????? mean?? ?6372510.886484
??? median?? ?6372511.224692
?????? min?? ?6372508.535736
????? rmse?? ?6372510.886484
?????? sse?? ?934004584962312.250000
?????? std?? ?0.971000

evo_ape tum gt.txt l.txt -as -p
同时用多个参数可以写在一起,比如“-a -s”写作“-as”。
$ evo_ape tum gt.txt l.txt -as -p
APE w.r.t. translation part (m)
(with Sim(3) Umeyama alignment)?????? max?? ?0.384438
????? mean?? ?0.156007
??? median?? ?0.141948
?????? min?? ?0.010909
????? rmse?? ?0.185972
?????? sse?? ?0.795467
?????? std?? ?0.101230


You can use Umeyama alignment as a pre-processing step:
??? --align or -a = SE(3) Umeyama alignment (rotation, translation)
??? --align --correct_scale or -as = Sim(3) Umeyama alignment (rotation, translation, scale)
??? --correct_scale or -s = scale alignmentScale or Sim(3) alignment is usually required for monocular SLAM, where you usually have random scale. SE(3) alignment is useful for the absolute pose error (evo_ape) if you want to measure the shape similarity of trajectories as best as possible. The alignment_demo.py script shows the different types of alignment with an example trajectory (as shown in the evo_traj documentation).
New since v1.5.0: A simple origin alignment that can be useful for drift/loop closure evaluation is available with --align_origin. This is not based on the Umeyama algorithm.
您可以使用Umeyama对齐作为预处理步骤:
--align或-a是SE(3)Umeyama对齐(旋转、平移)
--align—correct_scale或-as 是Sim(3)Umeyama对齐(旋转、平移、缩放)
--correct_scale或-s是缩放对齐
单目SLAM通常需要缩放或Sim(3)对齐,其中通常有随机缩放。如果你想尽可能最好地测量轨迹的形状相似性,SE(3)对齐对绝对姿态误差(evo_ape)很有用。alignment_demo.py脚本显示了具有示例轨迹的不同类型的对齐(如evo_traj文档中所示)。
v1.5.0以来的新增功能:--align_origin提供了一种简单的原点对齐,可用于漂移/环路闭合评估。这不是基于Umeyama算法。
evo_ape tum gt.txt l.txt? --n_to_align 1
? --n_to_align N_TO_ALIGN
??????????????????????? the number of poses to use for Umeyama alignment,counted from the start (default: all)用于Umeyama对齐的姿势数,从开始计数(默认值:全部)
$ evo_ape tum gt.txt l.txt? --n_to_align 1
APE w.r.t. translation part (m)
(not aligned)?????? max?? ?6372512.914678
????? mean?? ?6372511.473926
??? median?? ?6372511.871822
?????? min?? ?6372508.722174
????? rmse?? ?6372511.473926
?????? sse?? ?934004757162323.000000
?????? std?? ?1.138090
输出选项
evo_ape tum gt.txt l.txt -p
? -p, --plot??????????? show plot window显示绘图窗口
$ evo_ape tum gt.txt l.txt -p
APE w.r.t. translation part (m)
(not aligned)?????? max?? ?6372512.914678
????? mean?? ?6372511.473926
??? median?? ?6372511.871822
?????? min?? ?6372508.722174
????? rmse?? ?6372511.473926
?????? sse?? ?934004757162323.000000
?????? std?? ?1.138090

evo_ape tum gt.txt l.txt -as -p --plot_mode=xy
--plot_mode {xy,xz,yx,yz,zx,zy,xyz}
??????????????????????? the axes for plot projection绘图投影的轴
$ evo_ape tum gt.txt l.txt -as -p --plot_mode=xy
绘图投影在xy平面上。
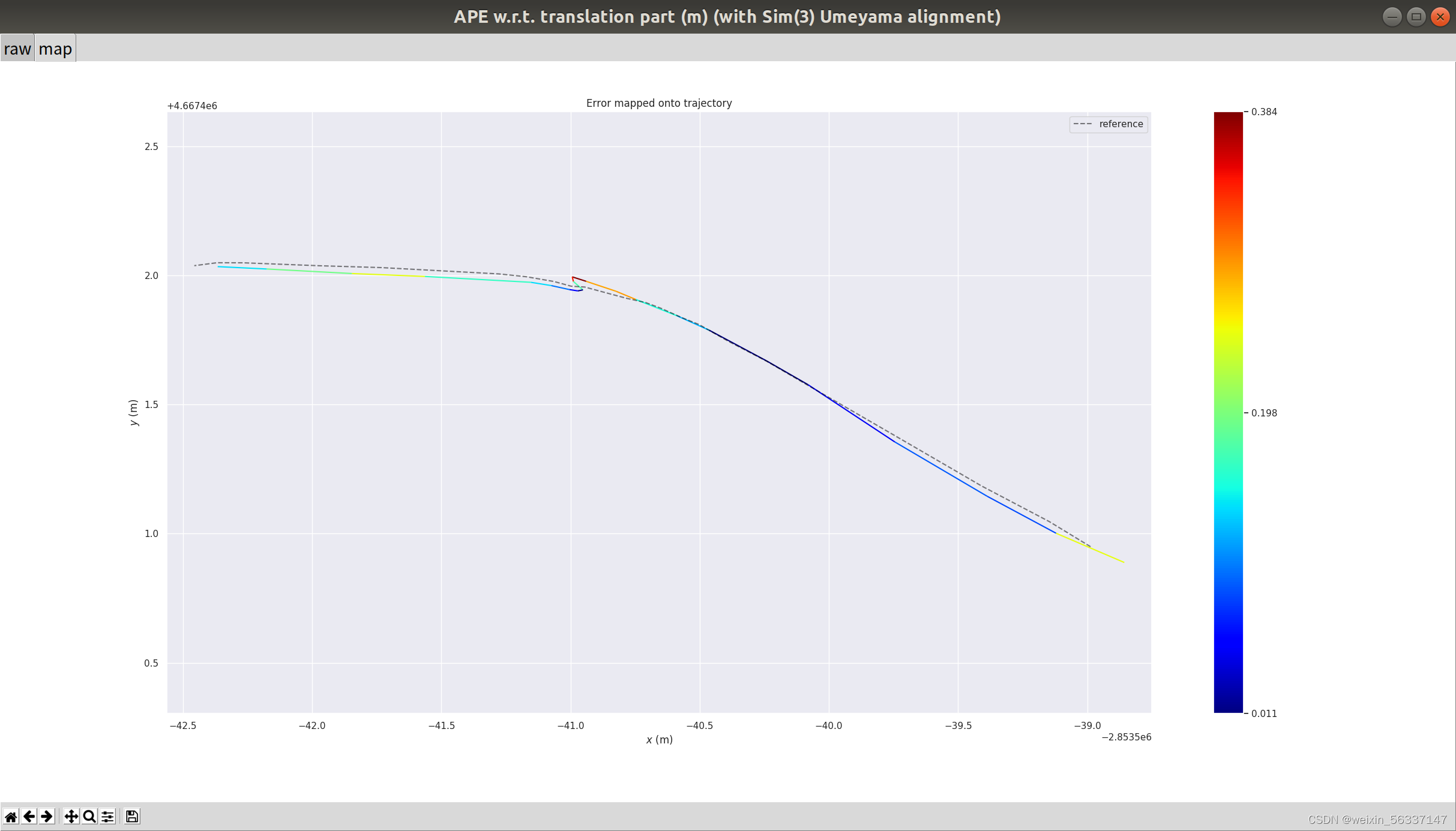
evo_ape tum gt.txt l.txt -as -p? --plot_colormap_max 0.2
? --plot_colormap_max PLOT_COLORMAP_MAX
??????????????????????? the upper bound used for the color map plot (default: maximum error value)用于颜色映射图的上限(默认值:最大误差值)
$ evo_ape tum gt.txt l.txt -as -p? --plot_colormap_max 0.2
颜色映射图的上限设为0.2,即0.2为红黑色。

evo_ape tum gt.txt l.txt -as -p --plot_colormap_min 0.2
? --plot_colormap_min PLOT_COLORMAP_MIN
??????????????????????? the lower bound used for the color map plot (default:minimum error value)用于颜色映射图的下限(默认值:最小误差值)
$ evo_ape tum gt.txt l.txt -as -p --plot_colormap_min 0.2
颜色映射图的下限设为0.2,即0.2为蓝黑色。

evo_ape tum gt.txt l.txt -as -p --plot_colormap_max_percentile 50
? --plot_colormap_max_percentile PLOT_COLORMAP_MAX_PERCENTILE
??????????????????????? percentile of the error distribution to be used as the? upper bound of the color map plot (in %, overrides? --plot_colormap_max)要用作颜色映射图上限的错误分布的百分位数(以%为单位,覆盖--plot_colormap_max)
$ evo_apeum gt.txt l.txt -as -p --plot_colormap_max_percentile 50
APE w.r.t. translation part (m)
(with Sim(3) Umeyama alignment)?????? max?? ?0.384438
????? mean?? ?0.156007
??? median?? ?0.141948
?????? min?? ?0.010909
????? rmse?? ?0.185972
?????? sse?? ?0.795467
?????? std?? ?0.101230
evo_ape tum gt.txt l.txt -as -p? --plot_full_ref
? --plot_full_ref?????? plot the full, unsynchronized reference trajectory绘制完整的、不同步的参考轨迹
$ evo_ape tum gt.txt l.txt -as -p? --plot_full_ref
(略)
我的理解是绘制轨迹没有平滑,存在原始数据的抖动。
evo_ape tum gt.txt l.txt -as? --save_plot SAVE_PLOT.pdf
? --save_plot SAVE_PLOT
??????????????????????? path to save plot保存绘图的路径
$ evo_ape tum gt.txt l.txt -as --save_plot SAVE_PLOT.pdf
(略)Plots saved to SAVE_PLOT
evo_ape tum gt.txt l.txt -as --serialize_plot SERIALIZE_PLOT
? --serialize_plot SERIALIZE_PLOT
??????????????????????? path to serialize plot (experimental)序列化绘图的路径(实验)
$ evo_ape tum gt.txt l.txt -as --serialize_plot SERIALIZE_PLOT
(略)
evo_ape tum gt.txt l.txt -as --save_results ape_result.zip
? --save_results SAVE_RESULTS
??????????????????????? .zip file path to store results存储结果的.zip文件路径
$ evo_ape tum gt.txt l.txt -as --save_results ape_result.zip
(略)
evo_ape tum gt.txt l.txt -as --logfile ape_result.log
? --logfile LOGFILE???? Local logfile path.本地日志文件路径。
$ evo_ape tum gt.txt l.txt -as --logfile ape_result.log
(略)
可用性选项
evo_ape tum gt.txt l.txt -as? --no_warnings
?--no_warnings???????? no warnings requiring user confirmation无需用户确认的警告
$ evo_ape tum gt.txt l.txt -as? --no_warnings
APE w.r.t. translation part (m)
(with Sim(3) Umeyama alignment)?????? max?? ?0.384438
????? mean?? ?0.156007
??? median?? ?0.141948
?????? min?? ?0.010909
????? rmse?? ?0.185972
?????? sse?? ?0.795467
?????? std?? ?0.101230
evo_ape tum gt.txt l.txt -v
? -v, --verbose???????? verbose output详细输出
$ evo_ape tum gt.txt l.txt -v
--------------------------------------------------------------------------------
Loaded 1684 stamps and poses from: gt.txt
Loaded 23 stamps and poses from: l.txt
Synchronizing trajectories...
Found 23 of max. 23 possible matching timestamps between...
?? ?gt.txt
and:?? ?l.txt
..with max. time diff.: 0.01 (s) and time offset: 0.0 (s).
--------------------------------------------------------------------------------
Compared 23 absolute pose pairs.
Calculating APE for translation part pose relation...
--------------------------------------------------------------------------------
APE w.r.t. translation part (m)
(not aligned)?????? max?? ?6372512.914678
????? mean?? ?6372511.473926
??? median?? ?6372511.871822
?????? min?? ?6372508.722174
????? rmse?? ?6372511.473926
?????? sse?? ?934004757162323.000000
?????? std?? ?1.138090
-v还可以搭配-s -a -sa等一起使用,主要是输出详细数据。
evo_ape tum gt.txt l.txt -as --silent
? --silent????????????? don't print any output不打印任何输出
$ evo_ape tum gt.txt l.txt -as --silent
?evo_ape tum gt.txt l.txt -as?? --debug
? --debug?????????????? verbose output with additional debug info使用附加调试信息调试详细输出
$ evo_ape tum gt.txt l.txt -as?? --debug
[DEBUG][2023-10-14 21:35:49,345][log.configure_logging():114]
System info:
Python 2.7.17
Linux-5.4.0-113-generic-x86_64-with-Ubuntu-18.04-bionic
?[DEBUG][2023-10-14 21:35:49,345][main_ape.run():227]
main_parser config:
{'align': True,
?'align_origin': False,
?'config': None,
?'correct_scale': True,
?'debug': True,
?'est_file': 'l.txt',
?'logfile': None,
?'n_to_align': -1,
?'no_warnings': False,
?'plot': False,
?'plot_colormap_max': None,
?'plot_colormap_max_percentile': None,
?'plot_colormap_min': None,
?'plot_full_ref': False,
?'plot_mode': u'xyz',
?'pose_relation': 'trans_part',
?'ref_file': 'gt.txt',
?'ros_map_yaml': None,
?'save_plot': None,
?'save_results': None,
?'serialize_plot': None,
?'silent': False,
?'subcommand': 'tum',
?'t_max_diff': 0.01,
?'t_offset': 0.0,
?'verbose': False}
[DEBUG][2023-10-14 21:35:49,345][main_ape.run():228]
--------------------------------------------------------------------------------
[DEBUG][2023-10-14 21:35:49,351][file_interface.read_tum_trajectory_file():112]
Loaded 1684 stamps and poses from: gt.txt
[DEBUG][2023-10-14 21:35:49,351][file_interface.read_tum_trajectory_file():112]
Loaded 23 stamps and poses from: l.txt
[DEBUG][2023-10-14 21:35:49,351][main_ape.run():238]
Synchronizing trajectories...
[DEBUG][2023-10-14 21:35:49,352][sync.associate_trajectories():105]
Found 23 of max. 23 possible matching timestamps between...
?? ?gt.txt
and:?? ?l.txt
..with max. time diff.: 0.01 (s) and time offset: 0.0 (s).
[DEBUG][2023-10-14 21:35:49,353][main_ape.ape():171]
--------------------------------------------------------------------------------
[DEBUG][2023-10-14 21:35:49,353][trajectory.align_trajectory():391]
Aligning using Umeyama's method... (with scale correction)
[DEBUG][2023-10-14 21:35:49,354][trajectory.align_trajectory():403]
Rotation of alignment:
[[-0.74522462 -0.51826266 -0.41957606]
?[ 0.03226245 -0.6565123?? 0.75362506]
?[-0.66603258? 0.5480834?? 0.50596955]]
Translation of alignment:
[-2853537.48321946? 4667400.58781658? 3268263.44928871]
[DEBUG][2023-10-14 21:35:49,354][trajectory.align_trajectory():404]
Scale correction: 0.849029484079
[DEBUG][2023-10-14 21:35:49,356][main_ape.ape():180]
--------------------------------------------------------------------------------
[DEBUG][2023-10-14 21:35:49,356][metrics.process_data():373]
Compared 23 absolute pose pairs.
[DEBUG][2023-10-14 21:35:49,356][metrics.process_data():375]
Calculating APE for translation part pose relation...
[DEBUG][2023-10-14 21:35:49,357][main_ape.ape():202]
--------------------------------------------------------------------------------
[INFO][2023-10-14 21:35:49,357][main_ape.ape():203]
APE w.r.t. translation part (m)
(with Sim(3) Umeyama alignment)?????? max?? ?0.384438
????? mean?? ?0.156007
??? median?? ?0.141948
?????? min?? ?0.010909
????? rmse?? ?0.185972
?????? sse?? ?0.795467
?????? std?? ?0.101230
本文来自互联网用户投稿,该文观点仅代表作者本人,不代表本站立场。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。 如若内容造成侵权/违法违规/事实不符,请联系我的编程经验分享网邮箱:veading@qq.com进行投诉反馈,一经查实,立即删除!